Workflow of G2S Web Service
The goal of G2S is to offer a high-accuracy, high-reliable, auto-updating computational services providing protein sequence to tertiary structure alignments in residue level. The workflow and architecture of G2S is shown as the following Figure. Solid lines are in Initiate workflow, dash lines are in Update workflow.
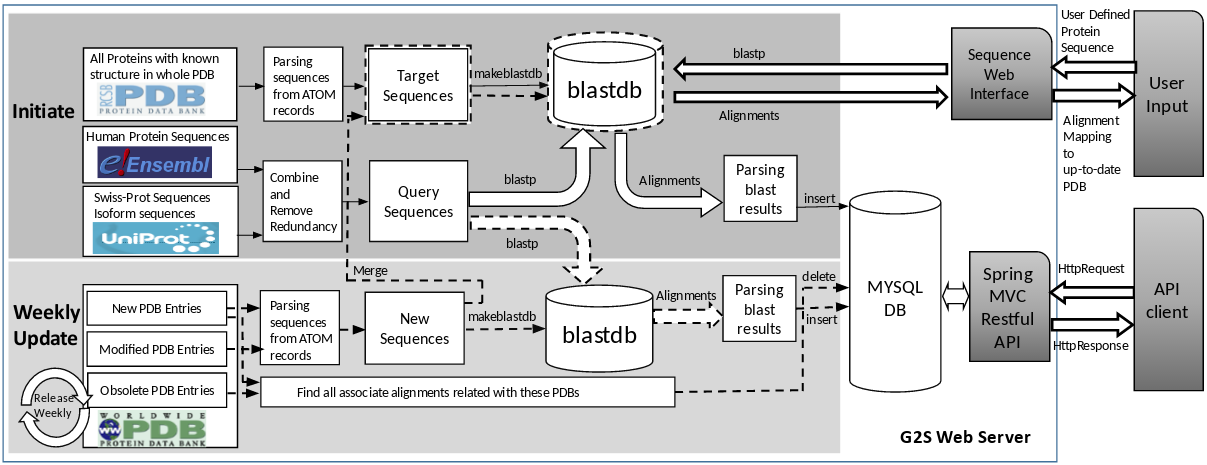
Figure 1: Initiate/Update Workflow and Architecture of G2S Web Service
To initiate the G2S web service, manually reviewed Uniprot (Swiss-Prot) sequences are integrated with Isoform sequences in recent release (2016_11) of UniProt (www.uniprot.org ), all sequences without isoforms are assigned as isoform 1. Then recent release of protein sequences from Human Release 87 in Ensembl (www.ensembl.org ) are integrated into UniProt sequences identified by the sequences. All identical sequences are combined together to remove redundancy. A strict threshold as 100% Redundancy is defined on the protein sequences, so only non-identical sequences will be left as an individual entry. Then these sequences are reorganized in internal SequenceId as the Query Sequences. The Target Sequences are elaborately parsing from ATOM records in PDB files from all solved protein tertiary structures in the whole PDB by BioJava (www.biojava.org ). Because of the uneven quality of PDB and technology limitations, chains of proteins often isolated by disorder regions failed to get structure solved. The solved pieces are re-sorted ascendingly and only the residues of the first biological assembly are chosen as the residue indices. To avoid too many small fragments, artificial repeat residues “X”s as Linker is used to connect these isolated pieces to keep continuity. Linked pieces failed to link with other pieces in a chain are defined as Segments. So segments in individual chain are organized as the basic unit of Target Sequences. In any Target Sequence, each residue in continuity has associated tertiary structure information comes from PDB. In G2S, the minimum length of Segment is set as 10, and the maximum Linker length is set as 5.
Once both Query and Target sequences are established, the Query Sequences are aligned with Target Sequences by BLASTP with predefined parameters. To avoid unnecessary non-informational results, each Query Sequence has maximum 50 alignments from Target Sequences sorted by Evalue. These alignment results are parsed and stored in a database. G2S API are implemented to obtain these pre-calculated alignments via querying this database by UniProt/Ensembl names for UniProt/Ensembl queries. Ensembl REST API is invoked to retrieve Ensembl names for the genomic region on Human Genome Version GRCH37. Besides adopt established protein names by the G2S API, any user-defined protein sequences could also be accepted and calculated in real time by the Sequence Web Interface. These input sequences will be aligned against up-to-date Target Sequences parsing form PDB files by BLASTP with user-defined parameters.
G2S has the capability of auto-updating of alignments from updating protein sequences against updating PDB. When PDB archive updates every Wednesday, the alignments of the obsolete and modified structures are abandoned, and only alignments between Query Sequences against new and modified structures in the PDB updates are inserted to the database. Meanwhile, these updates will be merged into Target Sequences. This update process is scheduled every week automatically to keep the database up to date with the PDB archive. When new version of UniProt and Ensembl released, the workflow will re-initiate and all Query Sequences will be regenerated, then the whole database will be replaced with new alignments between the Query Sequences and Target Sequences.